The Liau Lab conducts research at the interface of chemical biology and genomics. Our lab is collaborative and multidisciplinary, combining organic chemistry, cell and molecular biology, protein biochemistry, and large-scale genetic screening to dissect molecular mechanisms underpinning biological processes, cancer mutations, and drug mechanism of action. One major research focus is understanding protein complexes that control genome function, with the long-term goal of defining and targeting functional sites, mechanisms, and dependencies for small molecule development.
CHEMICAL GENOMICS
Our group embraces next-generation genome editing tools for chemical genetic approaches. In one major direction, we have advanced CRISPR mutational scanning approaches to interrogate protein structure-function and small molecule mechanism of action. These approaches enable the broad and systematic identification of functional mutations across endogenous proteins of interest. We study key mutations in deeper mechanistic studies using any method at our disposal, enabling us to deconvolute small molecule mechanism, uncover sites of protein functionality and allosteric regulation, and identify new cancer dependencies. We have expanded these approaches to investigate entire multi-subunit complexes and pathways, implement new forms of precision genome-editing and single-cell technologies, and explore the mechanisms of diverse small molecule modalities. Ongoing efforts are focused on exploring chromatin complexes, transcription factors, signaling molecules, and E3 ubiquitin ligases. Our long-term goal is to leverage large-scale chemical-genomic maps with mechanistic insight for the development of therapeutic approaches.
Publications:
- UM171 glues asymmetric CRL3-HDAC1/2 assembly to degrade CoREST corepressors. Yeo* et al. Nature Accepted.
- Converging mechanism of UM171 and KBTBD4 nomorphic cancer mutations. Xie* et al. Nature Accepted.
- Base editor scanning reveals activating mutations of DNMT3A. Garcia et al. ACS Chem Biol 2023.
- Drug addiction unveils a repressive methylation ceiling in EZH2-mutant lymphoma. Kwok* et al. Nat Chem Biol 2023.
- Activity-based CRISPR scanning uncovers allostery in DNA methylation maintenance machinery. Ngan et al. eLife 2023.
- Base editor scanning charts the DNMT3A activity landscape. Lue et al. Nat Chem Biol 2022.
- Profiling the landscape of drug resistance mutations in neosubstrates to molecular glue degraders. Gosavi* et al. ACS Cent Sci 2022.
- CRISPR-suppressor scanning reveals a nonenzymatic role of LSD1 in AML. Vinyard* et al. Nat Chem Biol 2019.
Reviews:
- Base editor screens for in situ mutational scanning at scale. Lue & Liau, Mol Cell 2023.
- Discovering new biology with drug-resistance alleles. Freedy & Liau, Nat Chem Biol 2021.
Gene Regulatory Complexes
We investigate the mechanisms of protein complexes that control genome function and transcription, using mutational scanning to identify their functional sites and chemical biology approaches to manipulate their activities and interactions. In particular, we have growing interests in studying transcription factors, their intertwined interactions with epigenetic regulators, and strategies to target their activities using small molecules.
Publications:
- An autoinhibitory switch of the LSD1 disordered region controls enhancer silencing. Waterbury* et al. Mol Cell 2024.
- DNA methylation insulates exons from CTCF loops near nuclear speckles. Roseman* et al. eLife 2024.
TECHNOLOGY DEVELOPMENT
We are developing molecular biology tools leveraged with next-generation sequencing to study different layers of gene regulation. To enable these and other efforts, we are interested in the chemical synthesis of small molecules and their functional derivatives.
Publications and Preprints:
- Coupling CRISPR scanning with targeted chromatin accessibility profiling using a double-stranded DNA deaminase. Roh*, Shen*, Hu* et al. bioRxiv 2024.
- Polycomb-lamina antagonism partitions heterochromatin at the nuclear periphery. Siegenfeld*, Roseman* et al. Nat. Commun 2022.
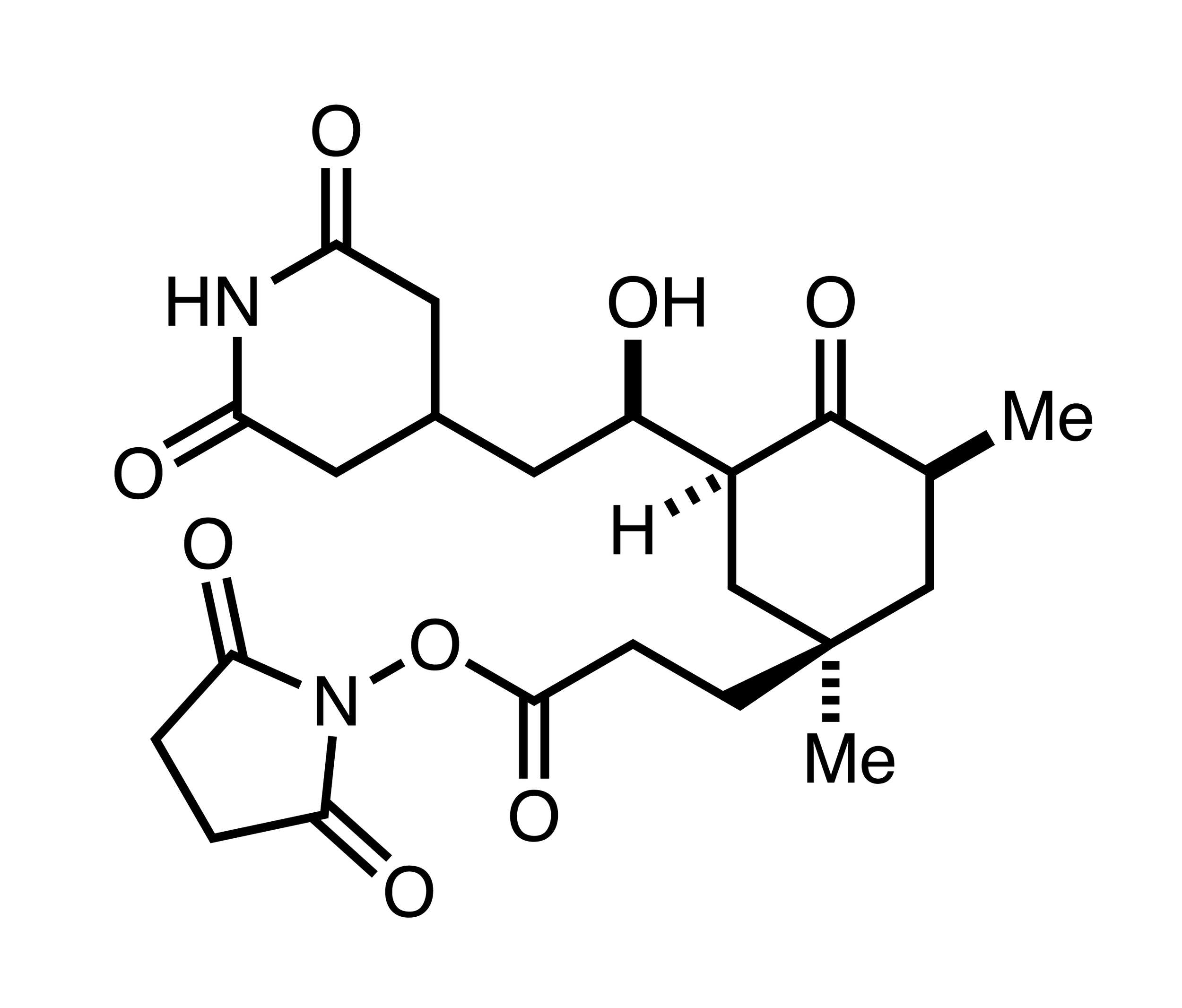
- Discovery of C13-aminobenzoyl cycloheximide derivatives that potently inhibit translation elongation. Koga et al. J Am Chem Soc 2021.
- A versatile synthetic route to cycloheximide and analogues that potently inhibit translation elongation. Park et al. Angew Chem Int Ed 2019.