Overview
We combine chemical biology with genetic perturbations at scale to study molecular mechanisms.
CHEMICAL GENOMICS
We study protein function and small molecule mechanism of action, particularly in the context of gene regulation. To do this, we are developing CRISPR mutational scanning approaches to systematically mutagenize target proteins directly in cells. We are expanding these approaches to investigate protein complexes and pathways at scale, where our long-term aim is to elucidate molecular mechanisms and therapeutic opportunities.
Publications and Preprints:
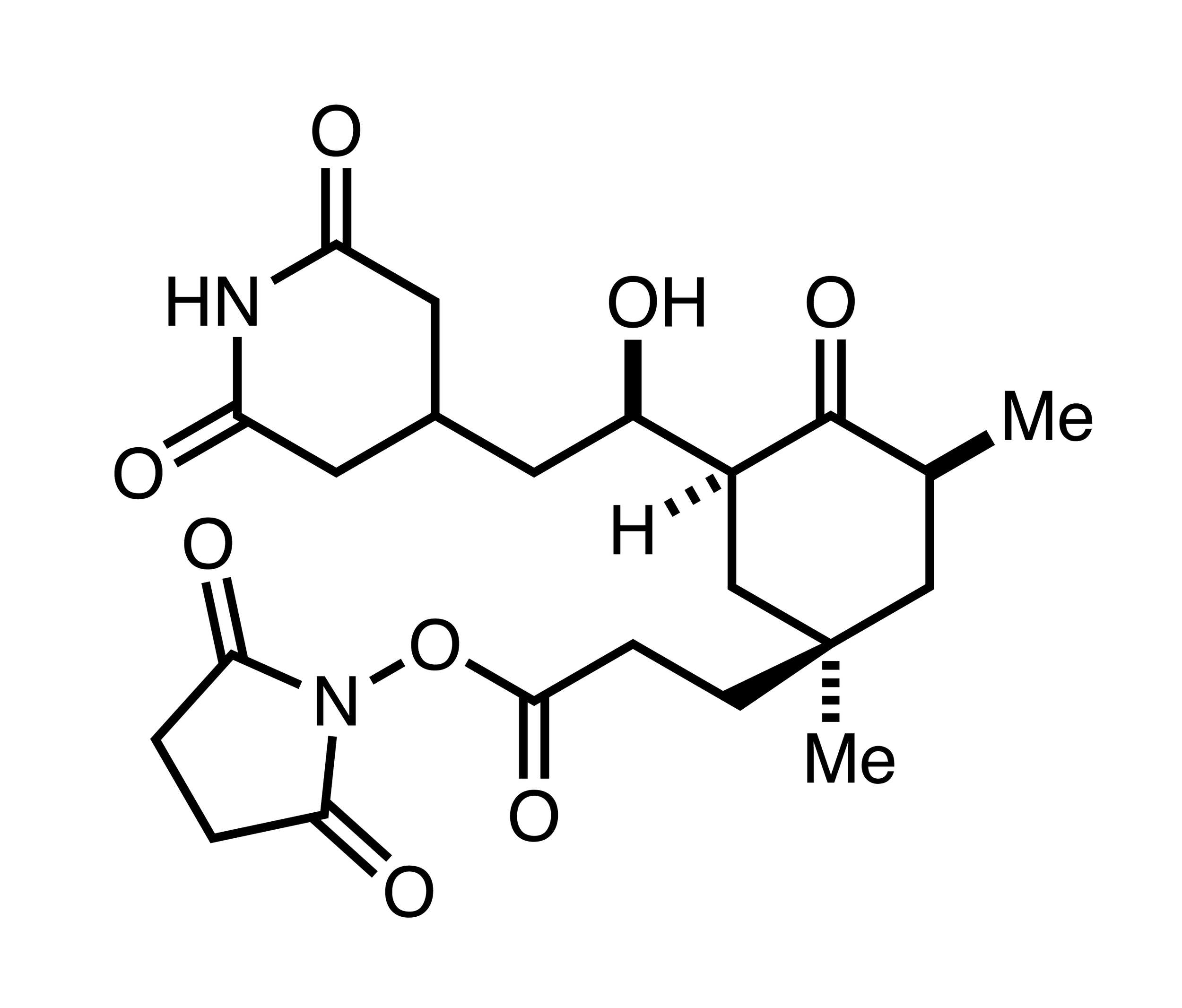
- UM171 glues asymmetric CRL3-HDAC1/2 assembly to degrade CoREST corepressors. Yeo et al. Nature (Accepted).
- Converging mechanism of UM171 and KBTBD4 neomorphic cancer mutations. Xie et al. Nature (Accepted).
- An autoinhibitory switch of the LSD1 disordered region controls enhancer silencing. Waterbury et al. Molecular Cell (2024).
- Base editor scanning reveals activating mutations of DNMT3A. Garcia et al. ACS Chemical Biology (2023).
- Drug addiction unveils a repressive methylation ceiling in EZH2-mutant lymphoma. Kwok et al. Nat. Chem. Biol. (2023).
- Activity-based CRISPR scanning uncovers allostery in DNA methylation maintenance machinery. Ngan et al. eLife (2023).
- Base editor scanning charts the DNMT3A activity landscape. Lue et al. Nat. Chem. Biol. (2023).
- Profiling the landscape of drug resistance mutations in neosubstrates to molecular glue degraders. Gosavi et al. ACS Cent. Sci. (2022).
- CRISPR-suppressor scanning reveals a nonenzymatic role of LSD1 in AML. Vinyard et al. Nat. Chem. Biol. (2019).
Technology Development
We are developing molecular biology tools leveraged with next-generation sequencing to study different layers of gene regulation. To enable these and other efforts, we are interested in the chemical synthesis of small molecules and their functional derivatives.
Publications and Preprints:
- Coupling CRISPR scanning with targeted chromatin accessibility profiling using a double-stranded DNA deaminase. Roh*, Shen*, Hu* et al. bioRxiv. (2024).
- Polycomb-lamina antagonism partitions heterochromatin at the nuclear periphery. Siegenfeld*, Roseman* et al. Nat. Commun. (2022).
- Discovery of C13-aminobenzoyl cycloheximide derivatives that potently inhibit translation elongation. Koga et al. J. Am. Chem. Soc. (2021).
- A versatile synthetic route to cycloheximide and analogues that potently inhibit translation elongation. Park et al. Angew. Chem. Int. Ed. (2019).